Abstract
Background: Philadelphia positive acute lymphoblastic leukaemia (Ph+ ALL) is a high risk ALL subtype with an aggressive clinical course. Despite significant improvements in outcomes combining tyrosine kinase inhibitors (TKIs) with chemotherapy, the development of resistance/relapse remains a major problem. Mutations in the phosphotyrosine phosphatase N-11 (PTPN11) gene are prevalent in haematological malignancies currently no targeted treatment options available clinically. In Ph+ ALL, it confers a poor prognosis, and may be implicated in treatment resistance against TKIs as well as emerging therapies such as venetoclax.
Aims: To investigate the role of PTPN11 mutations in the development of TKI resistance and to identify treatment strategies against those mutations in Ph+ ALL cells.
Methods: An imatinib resistant Ph+ ALL SUP-B15 line (resistant to 5 µM imatinib; SUP-B15 IR) was generated through incremental dose escalation of imatinib over 6 months. RNAseq and western blot analysis were performed to identify mechanisms of resistance. Short hairpin-RNAs specific for PTPN11 (encodes for SHP-2 protein) was introduced into SUP-B15 cells using lentiviral transduction and successfully transduced cells were selected by puromycin selection. Drug sensitivity was measured by Annexin V and viability stain 780. The combination index (CI) was calculated by Calcusyn software.
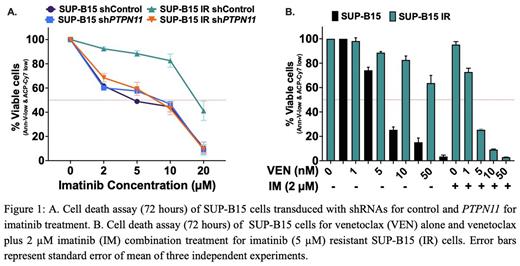
Results: The SUP-B15 IR cell line was found to have two PTPN11 mutations (p.A461T, VAF 64% and p.P491H, VAF 40%) from RNAseq analysis. Both of these mutations have previously been reported in Ph+ ALL patients. The SUP-B15 IR line was also resistant to nilotinib, dasatinib, ponatinib and asciminib as well as showing resistance to venetoclax (LD50 = 74.94 nM vs 2.1 nM in the parental, p=0.0054) (Figure 1B). SUP-B15 IRline showed no evidence of ABL1 kinase or myristoyl domain mutations. After genetically inhibiting (shRNA knockdown) PTPN11 in SUP-B15 IRcells, sensitivity to imatinib (Figure 1 A) and venetoclaxwas restored, confirming the central role of PTPN11 mutations in the development of TKI and venetoclax resistance in SUP-B15 IRcells. PTPN11 inhibition did not result in significant change in sensitivity of SUP-B15 parental cells to either imatinib or venetoclax treatment.
We looked for the most common mechanism of venetoclax resistance: the over-expression of other anti-apoptotic proteins such as MCL-1 and BCL-xL, and noted overexpression of MCL-1 (p<0.0001) and BCL-xL (p=0.0023) in SUP-B15 IR cells compared to the parental line. The overexpressed MCL-1 was mediated via an increase in pBCR::ABL1 (p<0.0001) shortly after imatinib removal in SUP-B15 IR cell line because both pBCR::ABL1 (p<0.0001) and MCL-1 (p<0.0001) were significantly inhibited with imatinib treatment but not with venetoclax treatmetent. However, BCL-xL expression was not affected by imatinib or venetoclax treatment suggesting BCL-xL may have compensated for the loss of MCL-1 when treated with imatinib and the loss of BCL-2 when treated with venetoclax. Treatment with imatinib also led to significant inhibition of pBCR::ABL1 (p<0.0001), but did not lead to reduction in MCL-1 expression in SUP-B15 parental cells suggesting modulation of MCL-1 expression via BCR::ABL1 activation was an acquired characteristic in SUP-B15 IRcells.
While the SUP-B15 IRcell line was also resistant to the MCL-1-selective inhibitor S63845 alone (LD50=290 nM vs 78.5 nM for parental), combination therapy using S63845 with venetoclax overcame the resistance (CI=0.053 for 5 nM venetoclax and 50 nM S63845). Interestingly, combination treatment with imatinib and venetoclax (CI = 0.049 for 2 µM IM and 5 nM VEN) (Figure 1B) or asciminib and venetoclax (CI=0.032 for 2 µM asciminib and 5 nM venetoclax) also overcame resistance in SUP-B15 IRcells.
Summary/Conclusions: Somatic mutations in PTPN11 in Ph+ ALL lead directly to TKI as well as venetoclax resistance rendering these mono-therapies ineffective. Combination therapy with TKIs plus venetoclax could be a promising treatment option for Ph+ ALL patients carrying PTPN11 mutations.
Disclosures
Yeung:Pfizer: Honoraria; Amgen: Honoraria; Takeda: Honoraria, Membership on an entity's Board of Directors or advisory committees; BMS: Honoraria, Research Funding; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding. White:BMS: Honoraria, Research Funding; Amgen: Honoraria. Hughes:Novartis: Consultancy, Research Funding; Enliven: Consultancy, Research Funding; BMS: Consultancy, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.